Sorting screen tutorial without reporter edits¶
Screens without editing outcome, where each gRNA is assigned to a target.
| Library design | Each guide RNA (or other perturbation such as ORF) has a specified target. |
|---|---|
| Selection | Cells are sorted based on FACS signal quantiles 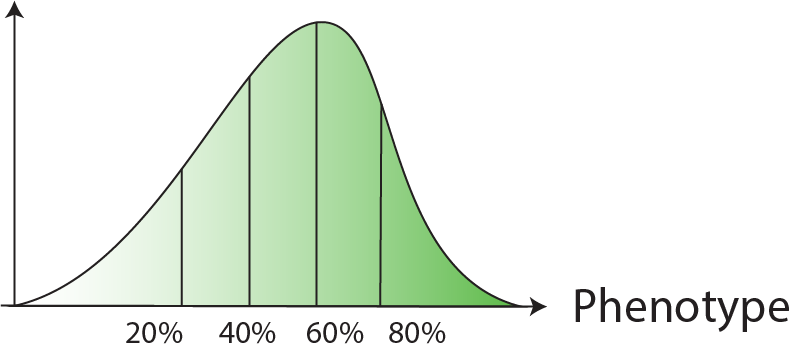 |
Here, we consider an example where BEAN uses the external count without reporter, gRNA and sample information to infer the per-target effect sizes.
Example workflow¶
screen_id=var_mini_screen_noedit
working_dir=tests/data/
# 1. Given that we have gRNA count for each sample, generate ReporterScreen (.h5ad) object for downstream analysis.
bean create-screen ${working_dir}/gRNA_info.csv ${working_dir}/sample_list.csv ${working_dir}/var_mini_counts.csv -o ${working_dir}/${screen_id}
# 2. QC samples & guides
bean qc \
${working_dir}/${screen_id}.h5ad `# Input ReporterScreen .h5ad file path` \
-o ${working_dir}/${screen_id}_masked.h5ad `# Output ReporterScreen .h5ad file path` \
-r ${working_dir}/qc_report_${screen_id} `# Prefix for QC report` \
-b ` # Remove replicates with no good samples.
# 3. Quantify variant effect
bean run sorting variant \
${working_dir}/${screen_id}_masked.h5ad \
-o ${working_dir}/ \
--uniform-edit --ignore-bcmatch `# As we have no edit/reporter information.` \
[--fit-negctrl [--negctrl-col target_group --negctrl-col-value NegCtrl]] `# If you have the negative control gRNAs.`
Input file spec¶
gRNA_info.csv: Should have
name,targetcolumns. You can also specifytarget_groupcolumn whose value indicatePosCtrl/NegCtrlfor control gRNAs.sample_list.csv: Same requirement for the full run. See examples for ``sorting` screens <https://github.com/pinellolab/crispr-bean/blob/main/tests/data/var_mini_samples.csv>`_ and ``survival` screens <https://github.com/pinellolab/crispr-bean/blob/main/tests/data/sample_list_survival.csv>`_.
See the example input files here.
See Subcommands for the full details.