bean profile¶
bean profile: Profile editing patterns¶
bean profile my_sorting_screen.h5ad -o output_prefix --pam-col '5-nt PAM'`# Prefix for editing profile report`
Output¶
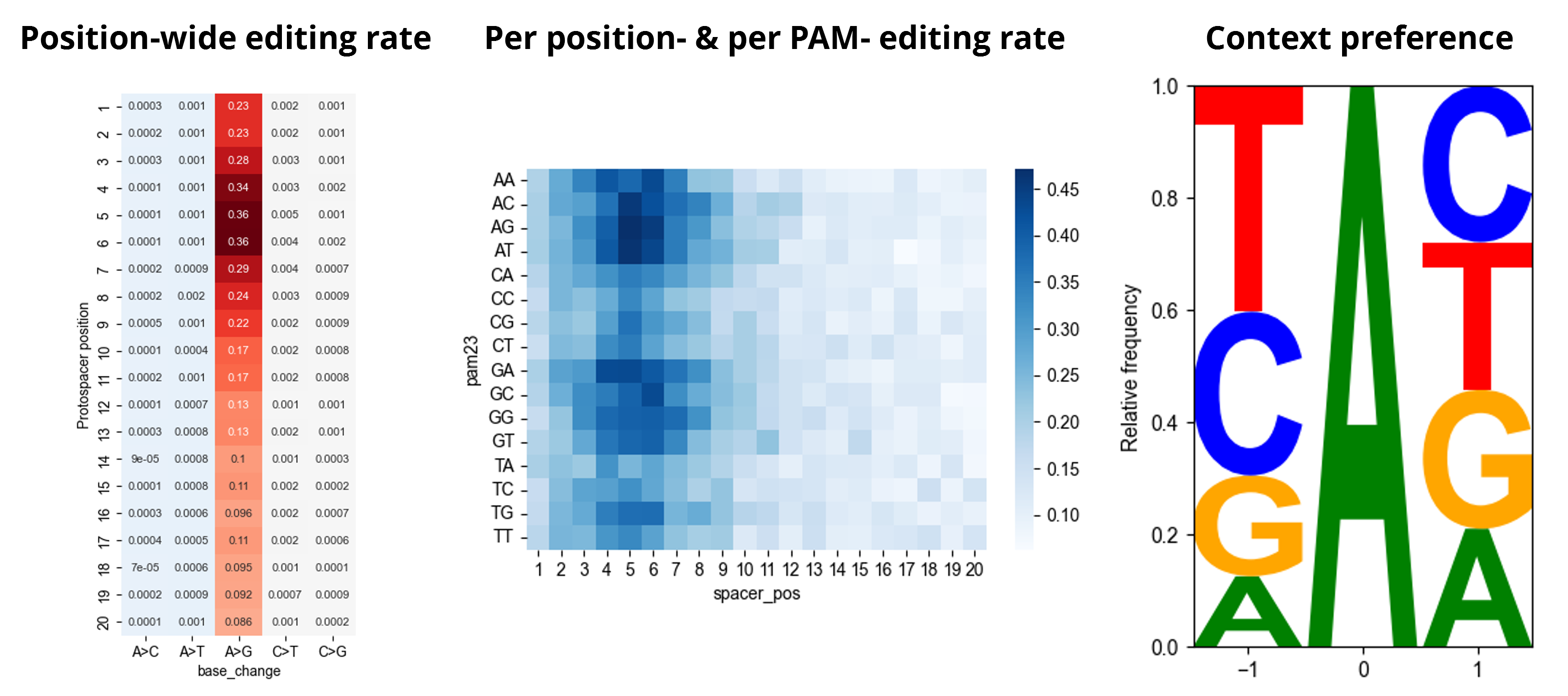
Above command produces prefix_editing_preference.[html,ipynb] as editing preferences (see example).
Full parameters¶
usage: bean profile [-h] [-o OUTPUT_PREFIX] [--replicate-col REPLICATE_COL]
[--condition-col CONDITION_COL] [--pam-col PAM_COL]
[--control-condition CONTROL_CONDITION] [-w WINDOW_LENGTH]
[--save-fig] [--reporter-length REPORTER_LENGTH]
[--reporter-right-flank-length REPORTER_RIGHT_FLANK_LENGTH]
bdata_path
Positional Arguments¶
- bdata_path
Path to the ReporterScreen object to run QC on
Named Arguments¶
- -o, --output-prefix
Output prefix of editing pattern report (prefix.html, prefix.ipynb). If not provided, base name of bdata_path is used.
- --replicate-col
Column name in bdata.samples that describes replicate ID.
Default:
'replicate'- --condition-col
Column name in bdata.samples that describes experimental condition. (sorting bin, time, etc.)
Default:
'condition'- --pam-col
Column name describing PAM of each gRNA in bdata.guides.
- --control-condition
Control condition where editing preference would be profiled at. Pre-filters data where bdata.samples[condition_col] == control_condition.
Default:
'bulk'- -w, --window-length
Window length of editing window of maximal editing efficiency to be identified. This window is used to quantify context specificity within the window.
Default:
6- --save-fig
Save .pdf of the figures included in the report.
Default:
False- --reporter-length
Length of reporter sequence in the construct.
Default:
32- --reporter-right-flank-length
Length of the right-flanking nucleotides of protospacer in the reporter.
Default:
6