PBMCs#
Load modules#
[18]:
%load_ext autoreload
%autoreload 2
import pyro
from mpl_toolkits.axes_grid1 import make_axes_locatable
import numpy as np
import matplotlib.pyplot as plt
from scipy.stats import pearsonr, spearmanr
import seaborn as sns
from pyrovelocity.utils import mRNA, debug, tau_inv, ode_mRNA
import torch
from torch import nn
import scvelo as scv
kwargs = dict(linewidth=1.5, density=.8, color='celltype', frameon=False, add_margin=.1, alpha=.1, min_mass=3.5, add_outline=True, outline_width=(.02, .02))
from scipy.stats import spearmanr, pearsonr
from pyrovelocity.api import train_model
import seaborn as sns
import pandas as pd
from pyrovelocity.plot import plot_posterior_time, plot_gene_ranking,\
vector_field_uncertainty, plot_vector_field_uncertain,\
plot_mean_vector_field, project_grid_points,rainbowplot,plot_arrow_examples
from pyrovelocity.utils import mae, mae_evaluate
The autoreload extension is already loaded. To reload it, use:
%reload_ext autoreload
Reproduce fully mature cell type problem#
[2]:
adata = scv.datasets.pbmc68k()
scv.pp.remove_duplicate_cells(adata)
adata.obsm["X_tsne"][:, 0] *= -1
[3]:
scv.pl.scatter(adata, c='celltype', s=30, legend_fontsize=12, legend_align_text=True, title='', figsize=(5, 5))
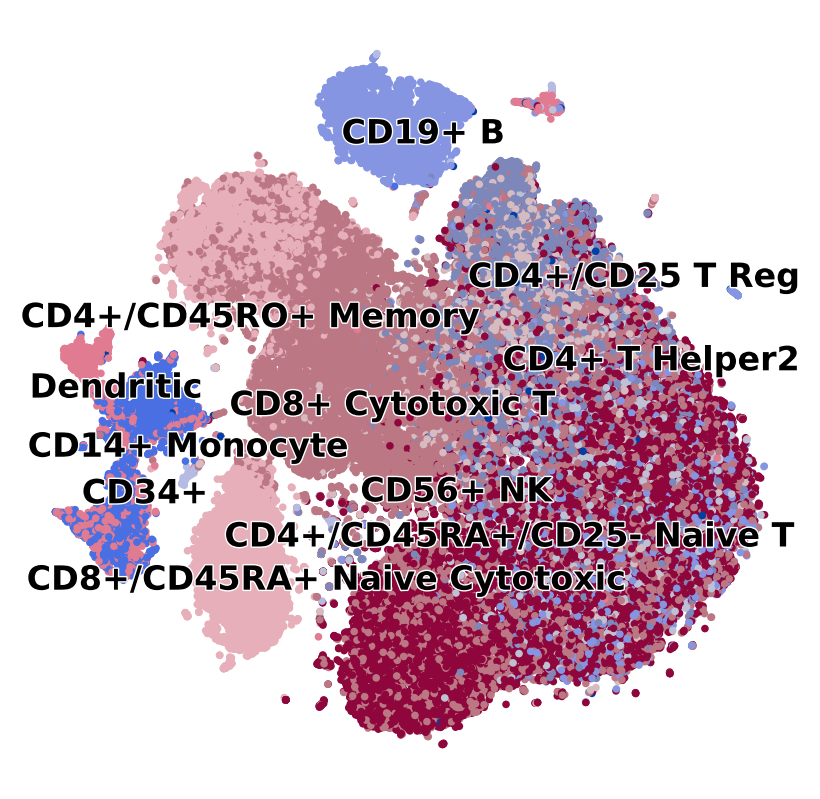
[4]:
scv.pp.filter_and_normalize(adata, min_shared_counts=30, n_top_genes=2000)
scv.pp.moments(adata)
Filtered out 33500 genes that are detected 30 counts (shared).
Normalized count data: X, spliced, unspliced.
Skip filtering by dispersion since number of variables are less than `n_top_genes`.
Logarithmized X.
computing neighbors
finished (0:00:38) --> added
'distances' and 'connectivities', weighted adjacency matrices (adata.obsp)
computing moments based on connectivities
finished (0:00:02) --> added
'Ms' and 'Mu', moments of un/spliced abundances (adata.layers)
[5]:
scv.tl.velocity(adata, mode='stochastic')
computing velocities
finished (0:00:02) --> added
'velocity', velocity vectors for each individual cell (adata.layers)
[6]:
scv.tl.recover_dynamics(adata, n_jobs=-1)
recovering dynamics (using 256/256 cores)
finished (0:00:46) --> added
'fit_pars', fitted parameters for splicing dynamics (adata.var)
[7]:
top_genes = adata.var['fit_likelihood'].sort_values(ascending=False).index
scv.pl.scatter(adata, basis=top_genes[:3], vkey='velocity', c='celltype', ncols=5, s=30, linewidth=2, frameon='artist', dpi=80)
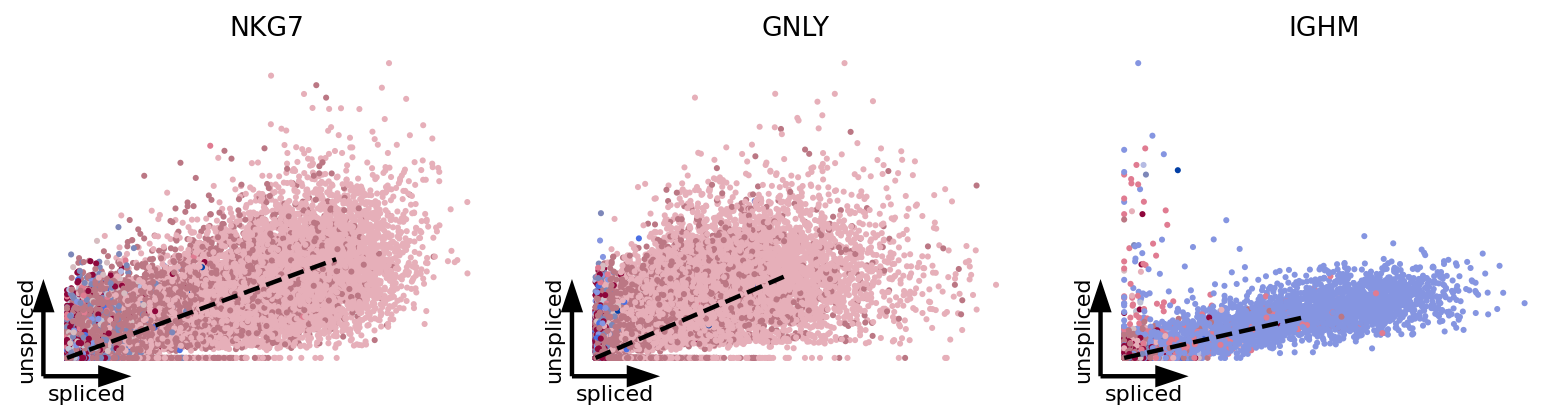
[8]:
adata_sub = adata[:, top_genes[:3]].copy()
[9]:
scv.tl.velocity_graph(adata_sub, n_jobs=-1)
scv.tl.velocity_embedding(adata_sub)
computing velocity graph (using 256/256 cores)
finished (0:00:55) --> added
'velocity_graph', sparse matrix with cosine correlations (adata.uns)
computing velocity embedding
finished (0:00:16) --> added
'velocity_tsne', embedded velocity vectors (adata.obsm)
[10]:
np.random.seed(99)
adata_sub.layers['velocity_with_noise'] = adata_sub.layers['velocity'] + np.random.normal(adata_sub.layers['velocity'], scale=adata_sub.layers['Ms'].std(0))
scv.tl.velocity_graph(adata_sub, gene_subset=top_genes[:3], vkey='velocity_with_noise', n_jobs=-1)
scv.tl.velocity_embedding(adata_sub, vkey='velocity_with_noise', autoscale=False)
computing velocity graph (using 256/256 cores)
finished (0:00:45) --> added
'velocity_with_noise_graph', sparse matrix with cosine correlations (adata.uns)
computing velocity embedding
finished (0:00:16) --> added
'velocity_with_noise_tsne', embedded velocity vectors (adata.obsm)
[11]:
gs = scv.pl.gridspec(2)
kwargs = dict(color='celltype', density=.8, add_margin=.1, alpha=.02, min_mass=3.5, add_outline=True, frameon=False)
scv.pl.velocity_embedding_stream(adata_sub, vkey='velocity_with_noise', legend_loc='none', title='expected (PBMC)', ax=gs[0], **kwargs)
scv.pl.velocity_embedding_stream(adata_sub, linewidth=1.5, legend_loc='right', title='estimated (PBMC)', ax=gs[1], **kwargs)
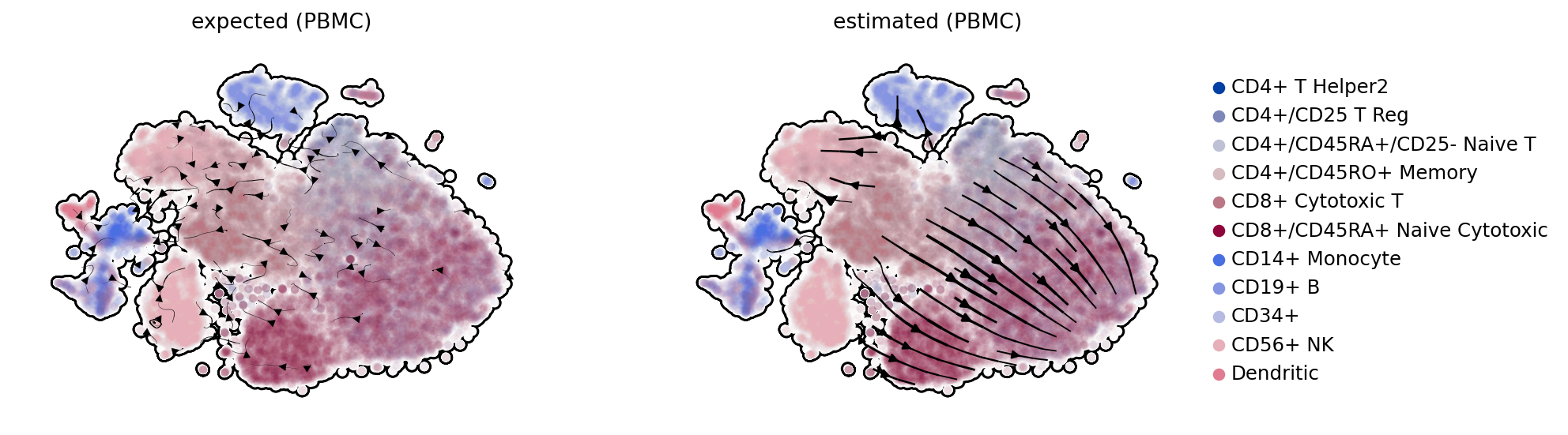
PBMC Pyro-velocity estimation of uncertainty#
[12]:
adata_all = scv.datasets.pbmc68k()
adata_all.obs['u_lib_size'] = adata_all.layers['unspliced'].toarray().sum(-1)
adata_all.obs['s_lib_size'] = adata_all.layers['spliced'].toarray().sum(-1)
adata_sub.layers['raw_spliced'] = adata_all[:, adata_sub.var_names].layers['spliced']
adata_sub.layers['raw_unspliced'] = adata_all[:, adata_sub.var_names].layers['unspliced']
adata_sub.obs['u_lib_size_raw'] = np.array(adata_sub.layers['raw_unspliced'].sum(axis=-1), dtype=np.float32).flatten()
adata_sub.obs['s_lib_size_raw'] = np.array(adata_sub.layers['raw_spliced'].sum(axis=-1), dtype=np.float32).flatten()
[15]:
adata_model_pos = train_model(adata_sub, max_epochs=4000, svi_train=False, log_every=100,
patient_init=45, batch_size=-1, use_gpu=1,
include_prior=True, offset=False, library_size=True,
cell_state='celltype',
patient_improve=1e-4, guide_type='auto_t0_constraint', train_size=0.67)
INFO No batch_key inputted, assuming all cells are same batch
INFO No label_key inputted, assuming all cells have same label
INFO Using data from adata.layers["raw_unspliced"]
INFO Using data from adata.layers["raw_spliced"]
INFO Successfully registered anndata object containing 65877 cells, 3 vars, 1 batches, 1 labels, and 0
proteins. Also registered 0 extra categorical covariates and 0 extra continuous covariates.
INFO Please do not further modify adata until model is trained.
Anndata setup with scvi-tools version 0.13.0.
Data Summary ┏━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━┳━━━━━━━┓ ┃ Data ┃ Count ┃ ┡━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━╇━━━━━━━┩ │ Cells │ 65877 │ │ Vars │ 3 │ │ Labels │ 1 │ │ Batches │ 1 │ │ Proteins │ 0 │ │ Extra Categorical Covariates │ 0 │ │ Extra Continuous Covariates │ 0 │ └──────────────────────────────┴───────┘
SCVI Data Registry ┏━━━━━━━━━━━━━━━━━━┳━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━┓ ┃ Data ┃ scvi-tools Location ┃ ┡━━━━━━━━━━━━━━━━━━╇━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━┩ │ X │ adata.layers['raw_spliced'] │ │ U │ adata.layers['raw_unspliced'] │ │ batch │ adata.obs['_scvi_batch'] │ │ label │ adata.obs['_scvi_labels'] │ │ ind_x │ adata.obs['_indices'] │ │ u_lib_size │ adata.obs['u_lib_size'] │ │ s_lib_size │ adata.obs['s_lib_size'] │ │ u_lib_size_mean │ adata.obs['u_lib_size_mean'] │ │ s_lib_size_mean │ adata.obs['s_lib_size_mean'] │ │ u_lib_size_scale │ adata.obs['u_lib_size_scale'] │ │ s_lib_size_scale │ adata.obs['s_lib_size_scale'] │ └──────────────────┴───────────────────────────────┘
Label Categories ┏━━━━━━━━━━━━━━━━━━━━━━━━━━━┳━━━━━━━━━━━━┳━━━━━━━━━━━━━━━━━━━━━┓ ┃ Source Location ┃ Categories ┃ scvi-tools Encoding ┃ ┡━━━━━━━━━━━━━━━━━━━━━━━━━━━╇━━━━━━━━━━━━╇━━━━━━━━━━━━━━━━━━━━━┩ │ adata.obs['_scvi_labels'] │ 0 │ 0 │ └───────────────────────────┴────────────┴─────────────────────┘
Batch Categories ┏━━━━━━━━━━━━━━━━━━━━━━━━━━┳━━━━━━━━━━━━┳━━━━━━━━━━━━━━━━━━━━━┓ ┃ Source Location ┃ Categories ┃ scvi-tools Encoding ┃ ┡━━━━━━━━━━━━━━━━━━━━━━━━━━╇━━━━━━━━━━━━╇━━━━━━━━━━━━━━━━━━━━━┩ │ adata.obs['_scvi_batch'] │ 0 │ 0 │ └──────────────────────────┴────────────┴─────────────────────┘
-----------
auto
auto_t0_constraint
step 0 loss = 1.74021e+06 patience = 45
step 100 loss = 1.28479e+06 patience = 45
step 200 loss = 1.01332e+06 patience = 45
step 300 loss = 848424 patience = 45
step 400 loss = 760072 patience = 45
step 500 loss = 702354 patience = 45
step 600 loss = 657546 patience = 43
step 700 loss = 638181 patience = 45
step 800 loss = 622194 patience = 45
step 900 loss = 593874 patience = 45
step 1000 loss = 587316 patience = 45
step 1100 loss = 568997 patience = 45
step 1200 loss = 650035 patience = 45
step 1300 loss = 568668 patience = 44
step 1400 loss = 592928 patience = 43
step 1500 loss = 1.6475e+06 patience = 45
step 1600 loss = 640139 patience = 44
step 1700 loss = 634879 patience = 45
step 1800 loss = 637467 patience = 44
step 1900 loss = 932600 patience = 45
step 2000 loss = 713719 patience = 45
step 2100 loss = 1.12977e+06 patience = 44
step 2200 loss = 769743 patience = 45
step 2300 loss = 894952 patience = 43
step 2400 loss = 1.14781e+06 patience = 45
step 2500 loss = 836674 patience = 45
step 2600 loss = 1.18126e+06 patience = 43
step 2700 loss = 1.10812e+06 patience = 45
step 2800 loss = 1.24692e+06 patience = 44
step 2900 loss = 922949 patience = 45
step 3000 loss = 2.57043e+06 patience = 45
step 3100 loss = 1.13558e+06 patience = 45
step 3200 loss = 1.05479e+06 patience = 45
step 3300 loss = 1.02177e+06 patience = 44
step 3400 loss = 1.15191e+06 patience = 45
step 3500 loss = 1.18264e+06 patience = 45
step 3600 loss = 900006 patience = 45
step 3700 loss = 982397 patience = 45
step 3800 loss = 546711 patience = 45
step 3900 loss = 609321 patience = 45
step 0 loss = 1.64668e+06 patience = 45
step 100 loss = 1.27097e+06 patience = 45
step 200 loss = 1.01447e+06 patience = 45
step 300 loss = 850348 patience = 45
step 400 loss = 756891 patience = 44
step 500 loss = 744063 patience = 45
step 600 loss = 1.10997e+06 patience = 45
step 700 loss = 1.16699e+06 patience = 45
step 800 loss = 1.51301e+06 patience = 43
step 900 loss = 2.33845e+06 patience = 43
step 1000 loss = 3.18e+07 patience = 44
step 1100 loss = 1.55246e+07 patience = 45
step 1200 loss = 8.10401e+06 patience = 44
step 1300 loss = 1.46762e+07 patience = 43
step 1400 loss = 4.14141e+07 patience = 45
step 1500 loss = 5.02087e+07 patience = 45
step 1600 loss = 1.99446e+08 patience = 45
step 1700 loss = 9.79143e+07 patience = 45
step 1800 loss = 8.20928e+07 patience = 44
step 1900 loss = 7.69921e+06 patience = 44
step 2000 loss = 3.35817e+07 patience = 44
step 2100 loss = 8.44306e+08 patience = 43
step 2200 loss = 7.807e+07 patience = 44
step 2300 loss = 8.31546e+07 patience = 45
step 2400 loss = 9.78603e+07 patience = 43
step 2500 loss = 6.14458e+07 patience = 45
step 2600 loss = 7.43112e+07 patience = 45
step 2700 loss = 1.01924e+07 patience = 43
step 2800 loss = 3.20883e+07 patience = 45
step 2900 loss = 8.38683e+07 patience = 45
step 3000 loss = 1.27062e+07 patience = 44
step 3100 loss = 1.83647e+07 patience = 44
step 3200 loss = 1.11801e+08 patience = 45
step 3300 loss = 6.62477e+08 patience = 45
step 3400 loss = 2.17665e+07 patience = 45
step 3500 loss = 3.37789e+07 patience = 45
step 3600 loss = 5.50768e+07 patience = 44
step 3700 loss = 3.06197e+07 patience = 44
step 3800 loss = 7.15672e+07 patience = 44
step 3900 loss = 4.65644e+07 patience = 45
[19]:
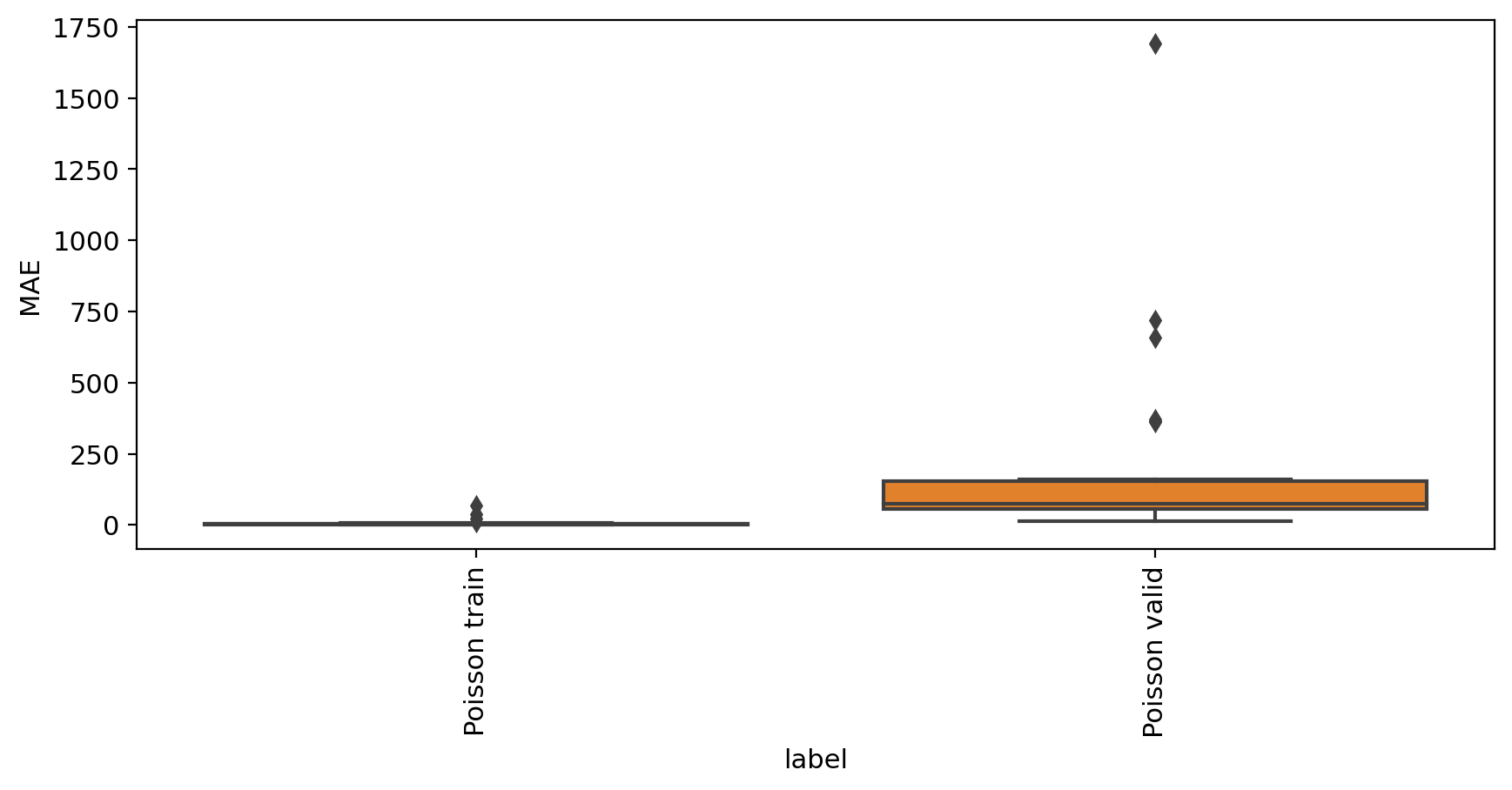
mae_evaluate(adata_model_pos, adata_sub)
MAE
label
Poisson train 6.980672
Poisson valid 200.731020
[19]:
| MAE | label | |
|---|---|---|
| 0 | 3.726239 | Poisson train |
| 1 | 2.510894 | Poisson train |
| 2 | 0.767772 | Poisson train |
| 3 | 1.719902 | Poisson train |
| 4 | 3.795361 | Poisson train |
| 5 | 2.484646 | Poisson train |
| 6 | 2.518356 | Poisson train |
| 7 | 2.931354 | Poisson train |
| 8 | 1.600094 | Poisson train |
| 9 | 1.509958 | Poisson train |
| 10 | 10.598647 | Poisson train |
| 11 | 1.846138 | Poisson train |
| 12 | 4.941194 | Poisson train |
| 13 | 0.829776 | Poisson train |
| 14 | 3.047674 | Poisson train |
| 15 | 2.685234 | Poisson train |
| 16 | 1.567151 | Poisson train |
| 17 | 1.851848 | Poisson train |
| 18 | 1.459392 | Poisson train |
| 19 | 2.350401 | Poisson train |
| 20 | 1.139426 | Poisson train |
| 21 | 1.113888 | Poisson train |
| 22 | 67.630816 | Poisson train |
| 23 | 5.277024 | Poisson train |
| 24 | 5.691608 | Poisson train |
| 25 | 38.824833 | Poisson train |
| 26 | 3.158665 | Poisson train |
| 27 | 3.161546 | Poisson train |
| 28 | 5.350277 | Poisson train |
| 29 | 23.330056 | Poisson train |
| 30 | 144.415823 | Poisson valid |
| 31 | 33.753596 | Poisson valid |
| 32 | 55.818890 | Poisson valid |
| 33 | 719.732965 | Poisson valid |
| 34 | 24.630136 | Poisson valid |
| 35 | 70.504400 | Poisson valid |
| 36 | 65.468215 | Poisson valid |
| 37 | 158.864643 | Poisson valid |
| 38 | 54.896220 | Poisson valid |
| 39 | 73.050330 | Poisson valid |
| 40 | 658.661638 | Poisson valid |
| 41 | 1690.662251 | Poisson valid |
| 42 | 361.695707 | Poisson valid |
| 43 | 22.728810 | Poisson valid |
| 44 | 73.196098 | Poisson valid |
| 45 | 150.909506 | Poisson valid |
| 46 | 14.789696 | Poisson valid |
| 47 | 74.471335 | Poisson valid |
| 48 | 12.422140 | Poisson valid |
| 49 | 147.348298 | Poisson valid |
| 50 | 36.116782 | Poisson valid |
| 51 | 365.234591 | Poisson valid |
| 52 | 63.321205 | Poisson valid |
| 53 | 40.944005 | Poisson valid |
| 54 | 86.226526 | Poisson valid |
| 55 | 371.670408 | Poisson valid |
| 56 | 153.898037 | Poisson valid |
| 57 | 104.048589 | Poisson valid |
| 58 | 57.008939 | Poisson valid |
| 59 | 135.440831 | Poisson valid |
[20]:
adata_model_pos = train_model(adata_sub, max_epochs=4000, svi_train=False, log_every=100,
patient_init=45, batch_size=-1, use_gpu=1, cell_state='celltype',
offset=False, library_size=True,
patient_improve=1e-4, guide_type='auto_t0_constraint', train_size=1.0,
include_prior=True)
INFO No batch_key inputted, assuming all cells are same batch
INFO:scvi.data._anndata:No batch_key inputted, assuming all cells are same batch
INFO No label_key inputted, assuming all cells have same label
INFO:scvi.data._anndata:No label_key inputted, assuming all cells have same label
INFO Using data from adata.layers["raw_unspliced"]
INFO:scvi.data._anndata:Using data from adata.layers["raw_unspliced"]
INFO Using data from adata.layers["raw_spliced"]
INFO:scvi.data._anndata:Using data from adata.layers["raw_spliced"]
INFO Successfully registered anndata object containing 65877 cells, 3 vars, 1 batches, 1 labels, and 0
proteins. Also registered 0 extra categorical covariates and 0 extra continuous covariates.
INFO:scvi.data._anndata:Successfully registered anndata object containing 65877 cells, 3 vars, 1 batches, 1 labels, and 0 proteins. Also registered 0 extra categorical covariates and 0 extra continuous covariates.
INFO Please do not further modify adata until model is trained.
INFO:scvi.data._anndata:Please do not further modify adata until model is trained.
Anndata setup with scvi-tools version 0.13.0.
Data Summary ┏━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━┳━━━━━━━┓ ┃ Data ┃ Count ┃ ┡━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━╇━━━━━━━┩ │ Cells │ 65877 │ │ Vars │ 3 │ │ Labels │ 1 │ │ Batches │ 1 │ │ Proteins │ 0 │ │ Extra Categorical Covariates │ 0 │ │ Extra Continuous Covariates │ 0 │ └──────────────────────────────┴───────┘
SCVI Data Registry ┏━━━━━━━━━━━━━━━━━━┳━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━┓ ┃ Data ┃ scvi-tools Location ┃ ┡━━━━━━━━━━━━━━━━━━╇━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━┩ │ X │ adata.layers['raw_spliced'] │ │ U │ adata.layers['raw_unspliced'] │ │ batch │ adata.obs['_scvi_batch'] │ │ label │ adata.obs['_scvi_labels'] │ │ ind_x │ adata.obs['_indices'] │ │ u_lib_size │ adata.obs['u_lib_size'] │ │ s_lib_size │ adata.obs['s_lib_size'] │ │ u_lib_size_mean │ adata.obs['u_lib_size_mean'] │ │ s_lib_size_mean │ adata.obs['s_lib_size_mean'] │ │ u_lib_size_scale │ adata.obs['u_lib_size_scale'] │ │ s_lib_size_scale │ adata.obs['s_lib_size_scale'] │ └──────────────────┴───────────────────────────────┘
Label Categories ┏━━━━━━━━━━━━━━━━━━━━━━━━━━━┳━━━━━━━━━━━━┳━━━━━━━━━━━━━━━━━━━━━┓ ┃ Source Location ┃ Categories ┃ scvi-tools Encoding ┃ ┡━━━━━━━━━━━━━━━━━━━━━━━━━━━╇━━━━━━━━━━━━╇━━━━━━━━━━━━━━━━━━━━━┩ │ adata.obs['_scvi_labels'] │ 0 │ 0 │ └───────────────────────────┴────────────┴─────────────────────┘
Batch Categories ┏━━━━━━━━━━━━━━━━━━━━━━━━━━┳━━━━━━━━━━━━┳━━━━━━━━━━━━━━━━━━━━━┓ ┃ Source Location ┃ Categories ┃ scvi-tools Encoding ┃ ┡━━━━━━━━━━━━━━━━━━━━━━━━━━╇━━━━━━━━━━━━╇━━━━━━━━━━━━━━━━━━━━━┩ │ adata.obs['_scvi_batch'] │ 0 │ 0 │ └──────────────────────────┴────────────┴─────────────────────┘
-----------
auto
auto_t0_constraint
TraceEnum
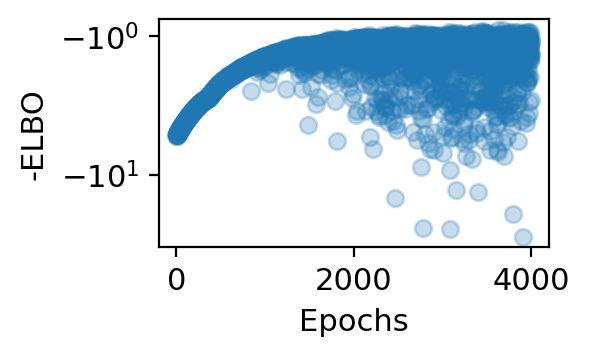
step 0 loss = 4.40379 patience = 45
step 100 loss = 3.25319 patience = 45
step 200 loss = 2.56925 patience = 45
step 300 loss = 2.15012 patience = 45
step 400 loss = 1.92119 patience = 45
step 500 loss = 1.76497 patience = 44
step 600 loss = 1.64497 patience = 44
step 700 loss = 1.55589 patience = 45
step 800 loss = 1.4728 patience = 45
step 900 loss = 1.40488 patience = 45
step 1000 loss = 1.34187 patience = 45
step 1100 loss = 1.29919 patience = 44
step 1200 loss = 1.25588 patience = 45
step 1300 loss = 1.19023 patience = 44
step 1400 loss = 1.21275 patience = 43
step 1500 loss = 1.53524 patience = 45
step 1600 loss = 1.14568 patience = 45
step 1700 loss = 1.11495 patience = 45
step 1800 loss = 1.07038 patience = 44
step 1900 loss = 1.18413 patience = 45
step 2000 loss = 1.06284 patience = 45
step 2100 loss = 1.15266 patience = 44
step 2200 loss = 1.11234 patience = 45
step 2300 loss = 1.15595 patience = 43
step 2400 loss = 1.20858 patience = 45
step 2500 loss = 1.08561 patience = 45
step 2600 loss = 1.23487 patience = 44
step 2700 loss = 1.215 patience = 41
step 2800 loss = 1.33119 patience = 44
step 2900 loss = 1.0495 patience = 45
step 3000 loss = 1.55312 patience = 45
step 3100 loss = 1.21313 patience = 44
step 3200 loss = 1.14989 patience = 45
step 3300 loss = 1.20562 patience = 43
step 3400 loss = 1.13651 patience = 43
step 3500 loss = 1.07121 patience = 44
step 3600 loss = 1.07778 patience = 45
step 3700 loss = 1.11148 patience = 45
step 3800 loss = 0.941839 patience = 45
step 3900 loss = 0.98063 patience = 45
[22]:
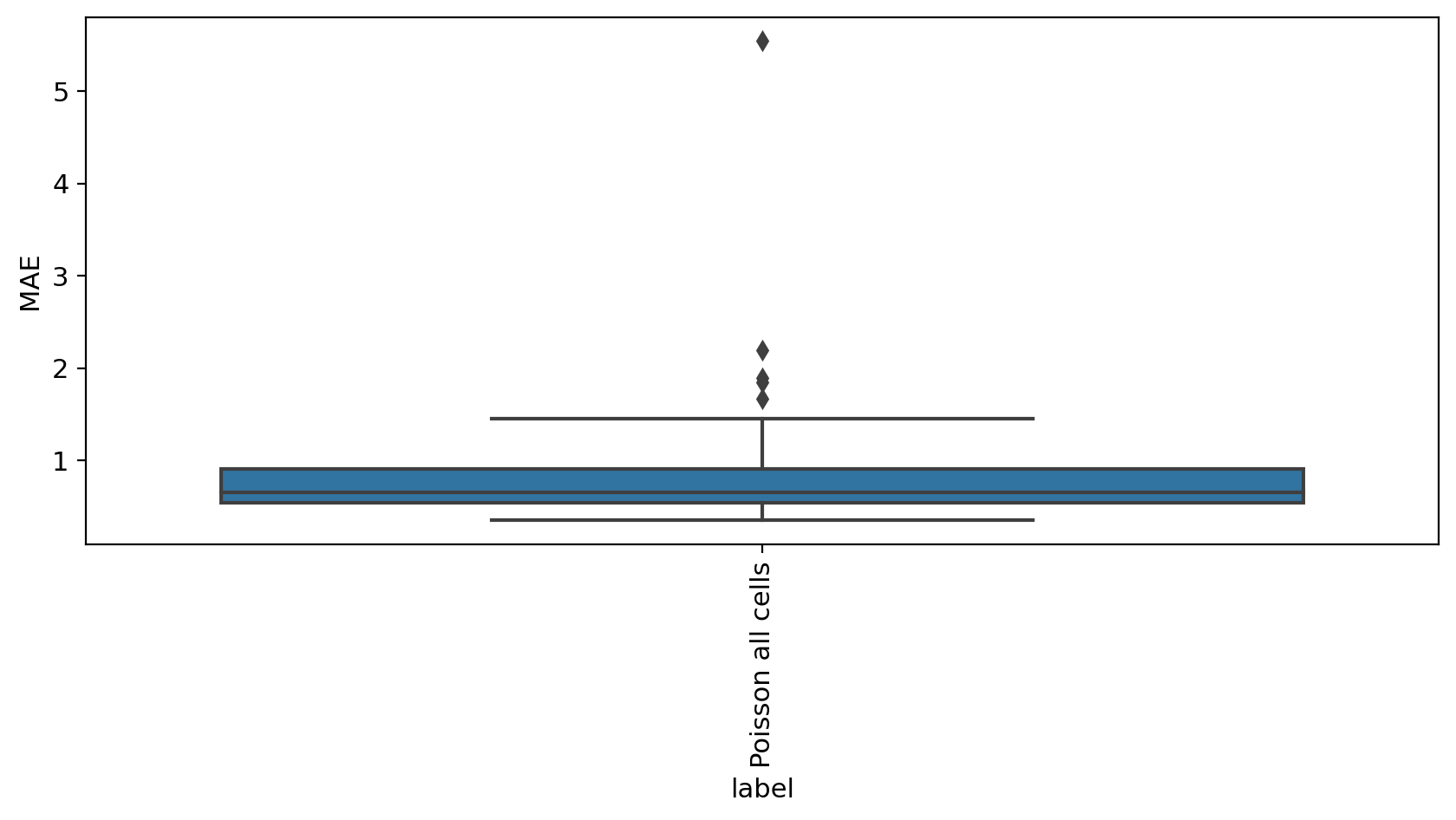
mae_evaluate(adata_model_pos[1], adata_sub)
MAE
label
Poisson all cells 1.002032
[22]:
| MAE | label | |
|---|---|---|
| 0 | 0.656059 | Poisson all cells |
| 1 | 0.640846 | Poisson all cells |
| 2 | 0.351339 | Poisson all cells |
| 3 | 1.448229 | Poisson all cells |
| 4 | 0.693919 | Poisson all cells |
| 5 | 1.402847 | Poisson all cells |
| 6 | 0.561997 | Poisson all cells |
| 7 | 0.538951 | Poisson all cells |
| 8 | 0.525482 | Poisson all cells |
| 9 | 0.540181 | Poisson all cells |
| 10 | 1.671800 | Poisson all cells |
| 11 | 0.588668 | Poisson all cells |
| 12 | 0.758687 | Poisson all cells |
| 13 | 0.545345 | Poisson all cells |
| 14 | 0.652587 | Poisson all cells |
| 15 | 1.848784 | Poisson all cells |
| 16 | 0.418606 | Poisson all cells |
| 17 | 0.639803 | Poisson all cells |
| 18 | 0.622068 | Poisson all cells |
| 19 | 0.530395 | Poisson all cells |
| 20 | 0.427220 | Poisson all cells |
| 21 | 0.373909 | Poisson all cells |
| 22 | 5.542068 | Poisson all cells |
| 23 | 0.924116 | Poisson all cells |
| 24 | 0.865340 | Poisson all cells |
| 25 | 2.196839 | Poisson all cells |
| 26 | 0.692958 | Poisson all cells |
| 27 | 0.685500 | Poisson all cells |
| 28 | 0.819634 | Poisson all cells |
| 29 | 1.896777 | Poisson all cells |
[24]:
v_map_all, embeds_radian, fdri = vector_field_uncertainty(adata_sub, adata_model_pos[1], n_jobs=20)
WARNING: You seem to have 414 duplicate cells in your data. Consider removing these via pp.remove_duplicate_cells.
computing neighbors
finished (0:00:18) --> added
'distances' and 'connectivities', weighted adjacency matrices (adata.obsp)
computing velocity graph (using 20/256 cores)
finished (0:00:56) --> added
'velocity_pyro_graph', sparse matrix with cosine correlations (adata.uns)
computing velocity embedding
finished (0:00:17) --> added
'velocity_pyro_tsne', embedded velocity vectors (adata.obsm)
computing velocity graph (using 20/256 cores)
finished (0:01:00) --> added
'velocity_pyro_graph', sparse matrix with cosine correlations (adata.uns)
computing velocity embedding
finished (0:00:17) --> added
'velocity_pyro_tsne', embedded velocity vectors (adata.obsm)
computing velocity graph (using 20/256 cores)
finished (0:00:53) --> added
'velocity_pyro_graph', sparse matrix with cosine correlations (adata.uns)
computing velocity embedding
finished (0:00:17) --> added
'velocity_pyro_tsne', embedded velocity vectors (adata.obsm)
computing velocity graph (using 20/256 cores)
finished (0:01:02) --> added
'velocity_pyro_graph', sparse matrix with cosine correlations (adata.uns)
computing velocity embedding
finished (0:00:16) --> added
'velocity_pyro_tsne', embedded velocity vectors (adata.obsm)
computing velocity graph (using 20/256 cores)
finished (0:00:58) --> added
'velocity_pyro_graph', sparse matrix with cosine correlations (adata.uns)
computing velocity embedding
finished (0:00:16) --> added
'velocity_pyro_tsne', embedded velocity vectors (adata.obsm)
computing velocity graph (using 20/256 cores)
finished (0:00:54) --> added
'velocity_pyro_graph', sparse matrix with cosine correlations (adata.uns)
computing velocity embedding
finished (0:00:16) --> added
'velocity_pyro_tsne', embedded velocity vectors (adata.obsm)
computing velocity graph (using 20/256 cores)
finished (0:00:59) --> added
'velocity_pyro_graph', sparse matrix with cosine correlations (adata.uns)
computing velocity embedding
finished (0:00:16) --> added
'velocity_pyro_tsne', embedded velocity vectors (adata.obsm)
computing velocity graph (using 20/256 cores)
finished (0:00:52) --> added
'velocity_pyro_graph', sparse matrix with cosine correlations (adata.uns)
computing velocity embedding
finished (0:00:16) --> added
'velocity_pyro_tsne', embedded velocity vectors (adata.obsm)
computing velocity graph (using 20/256 cores)
finished (0:01:00) --> added
'velocity_pyro_graph', sparse matrix with cosine correlations (adata.uns)
computing velocity embedding
finished (0:00:16) --> added
'velocity_pyro_tsne', embedded velocity vectors (adata.obsm)
computing velocity graph (using 20/256 cores)
finished (0:00:53) --> added
'velocity_pyro_graph', sparse matrix with cosine correlations (adata.uns)
computing velocity embedding
finished (0:00:16) --> added
'velocity_pyro_tsne', embedded velocity vectors (adata.obsm)
computing velocity graph (using 20/256 cores)
finished (0:01:02) --> added
'velocity_pyro_graph', sparse matrix with cosine correlations (adata.uns)
computing velocity embedding
finished (0:00:16) --> added
'velocity_pyro_tsne', embedded velocity vectors (adata.obsm)
computing velocity graph (using 20/256 cores)
finished (0:00:54) --> added
'velocity_pyro_graph', sparse matrix with cosine correlations (adata.uns)
computing velocity embedding
finished (0:00:16) --> added
'velocity_pyro_tsne', embedded velocity vectors (adata.obsm)
computing velocity graph (using 20/256 cores)
finished (0:01:00) --> added
'velocity_pyro_graph', sparse matrix with cosine correlations (adata.uns)
computing velocity embedding
finished (0:00:16) --> added
'velocity_pyro_tsne', embedded velocity vectors (adata.obsm)
computing velocity graph (using 20/256 cores)
finished (0:00:55) --> added
'velocity_pyro_graph', sparse matrix with cosine correlations (adata.uns)
computing velocity embedding
finished (0:00:16) --> added
'velocity_pyro_tsne', embedded velocity vectors (adata.obsm)
computing velocity graph (using 20/256 cores)
finished (0:01:02) --> added
'velocity_pyro_graph', sparse matrix with cosine correlations (adata.uns)
computing velocity embedding
finished (0:00:16) --> added
'velocity_pyro_tsne', embedded velocity vectors (adata.obsm)
computing velocity graph (using 20/256 cores)
finished (0:00:54) --> added
'velocity_pyro_graph', sparse matrix with cosine correlations (adata.uns)
computing velocity embedding
finished (0:00:16) --> added
'velocity_pyro_tsne', embedded velocity vectors (adata.obsm)
computing velocity graph (using 20/256 cores)
finished (0:01:02) --> added
'velocity_pyro_graph', sparse matrix with cosine correlations (adata.uns)
computing velocity embedding
finished (0:00:16) --> added
'velocity_pyro_tsne', embedded velocity vectors (adata.obsm)
computing velocity graph (using 20/256 cores)
finished (0:00:59) --> added
'velocity_pyro_graph', sparse matrix with cosine correlations (adata.uns)
computing velocity embedding
finished (0:00:16) --> added
'velocity_pyro_tsne', embedded velocity vectors (adata.obsm)
computing velocity graph (using 20/256 cores)
finished (0:00:54) --> added
'velocity_pyro_graph', sparse matrix with cosine correlations (adata.uns)
computing velocity embedding
finished (0:00:16) --> added
'velocity_pyro_tsne', embedded velocity vectors (adata.obsm)
computing velocity graph (using 20/256 cores)
finished (0:00:59) --> added
'velocity_pyro_graph', sparse matrix with cosine correlations (adata.uns)
computing velocity embedding
finished (0:00:16) --> added
'velocity_pyro_tsne', embedded velocity vectors (adata.obsm)
computing velocity graph (using 20/256 cores)
finished (0:00:53) --> added
'velocity_pyro_graph', sparse matrix with cosine correlations (adata.uns)
computing velocity embedding
finished (0:00:16) --> added
'velocity_pyro_tsne', embedded velocity vectors (adata.obsm)
computing velocity graph (using 20/256 cores)
finished (0:00:59) --> added
'velocity_pyro_graph', sparse matrix with cosine correlations (adata.uns)
computing velocity embedding
finished (0:00:16) --> added
'velocity_pyro_tsne', embedded velocity vectors (adata.obsm)
computing velocity graph (using 20/256 cores)
finished (0:00:53) --> added
'velocity_pyro_graph', sparse matrix with cosine correlations (adata.uns)
computing velocity embedding
finished (0:00:16) --> added
'velocity_pyro_tsne', embedded velocity vectors (adata.obsm)
computing velocity graph (using 20/256 cores)
finished (0:01:05) --> added
'velocity_pyro_graph', sparse matrix with cosine correlations (adata.uns)
computing velocity embedding
finished (0:00:16) --> added
'velocity_pyro_tsne', embedded velocity vectors (adata.obsm)
computing velocity graph (using 20/256 cores)
finished (0:00:53) --> added
'velocity_pyro_graph', sparse matrix with cosine correlations (adata.uns)
computing velocity embedding
finished (0:00:16) --> added
'velocity_pyro_tsne', embedded velocity vectors (adata.obsm)
computing velocity graph (using 20/256 cores)
finished (0:01:02) --> added
'velocity_pyro_graph', sparse matrix with cosine correlations (adata.uns)
computing velocity embedding
finished (0:00:17) --> added
'velocity_pyro_tsne', embedded velocity vectors (adata.obsm)
computing velocity graph (using 20/256 cores)
finished (0:00:54) --> added
'velocity_pyro_graph', sparse matrix with cosine correlations (adata.uns)
computing velocity embedding
finished (0:00:16) --> added
'velocity_pyro_tsne', embedded velocity vectors (adata.obsm)
computing velocity graph (using 20/256 cores)
finished (0:01:00) --> added
'velocity_pyro_graph', sparse matrix with cosine correlations (adata.uns)
computing velocity embedding
finished (0:00:16) --> added
'velocity_pyro_tsne', embedded velocity vectors (adata.obsm)
computing velocity graph (using 20/256 cores)
finished (0:00:54) --> added
'velocity_pyro_graph', sparse matrix with cosine correlations (adata.uns)
computing velocity embedding
finished (0:00:16) --> added
'velocity_pyro_tsne', embedded velocity vectors (adata.obsm)
computing velocity graph (using 20/256 cores)
finished (0:00:59) --> added
'velocity_pyro_graph', sparse matrix with cosine correlations (adata.uns)
computing velocity embedding
finished (0:00:16) --> added
'velocity_pyro_tsne', embedded velocity vectors (adata.obsm)
[26]:
fig, ax = plt.subplots()
embed_mean = plot_mean_vector_field(adata_model_pos[1], adata_sub, ax=ax, basis='tsne', n_jobs=20)
WARNING: You seem to have 414 duplicate cells in your data. Consider removing these via pp.remove_duplicate_cells.
computing neighbors
finished (0:00:18) --> added
'distances' and 'connectivities', weighted adjacency matrices (adata.obsp)
computing velocity graph (using 20/256 cores)
finished (0:01:04) --> added
'velocity_pyro_graph', sparse matrix with cosine correlations (adata.uns)
computing velocity embedding
finished (0:00:17) --> added
'velocity_pyro_tsne', embedded velocity vectors (adata.obsm)
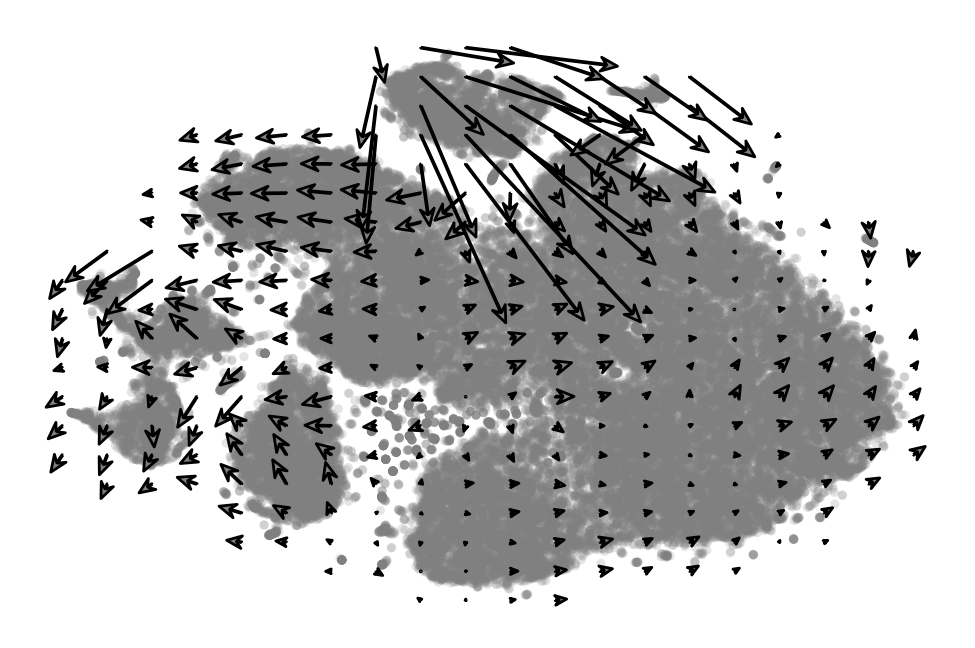
[27]:
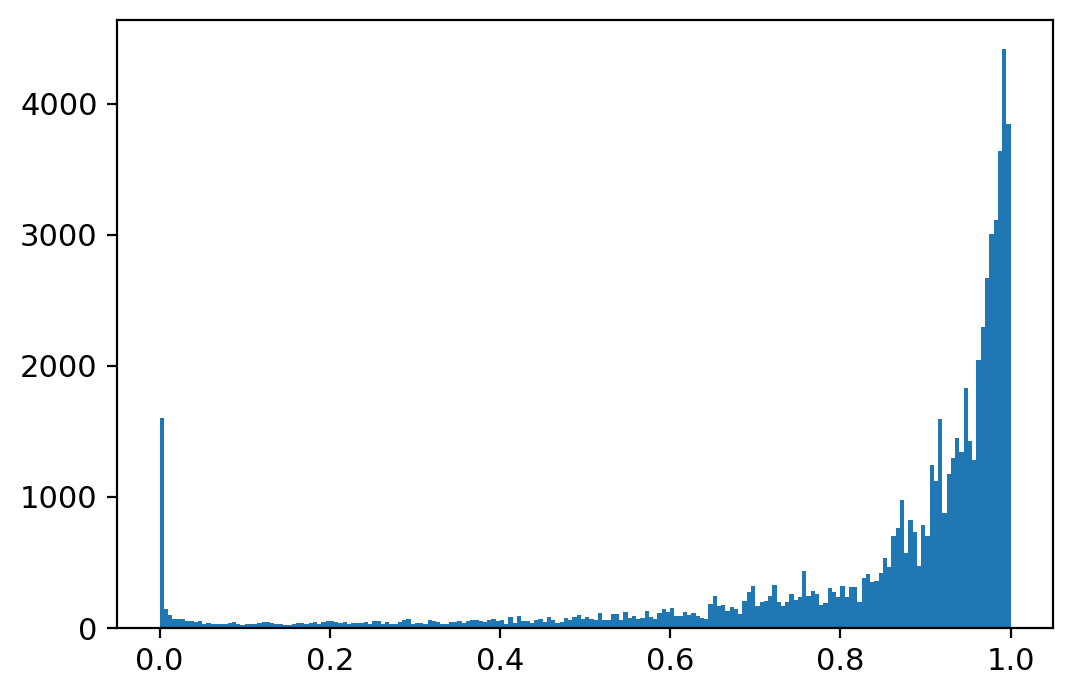
(fdri < 0.01).sum(), (fdri < 0.01).sum()/fdri.shape[0], fdri.shape, adata_sub.shape
[27]:
(1743, 0.02645839974497928, (65877,), (65877, 3))
[28]:
_ = plt.hist(fdri, bins=200)
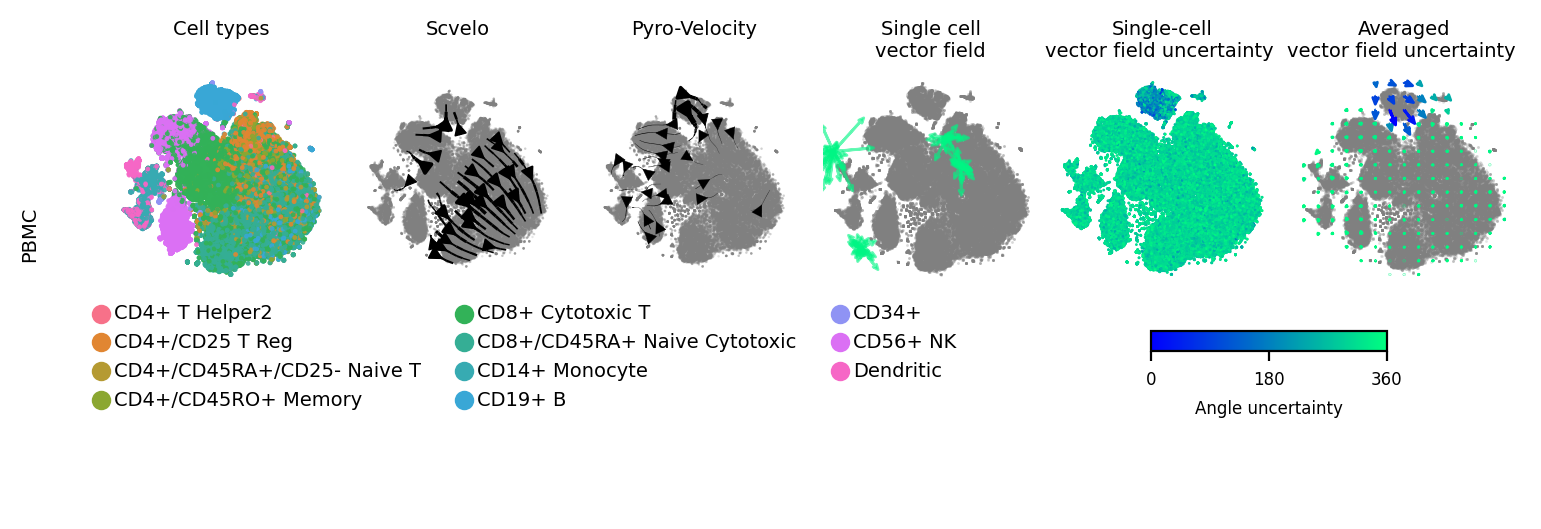
[34]:
fig = plt.figure(figsize=(7.07, 2.2))
subfig_A = fig.subfigures(2, 1, wspace=0.0, hspace=0, height_ratios=[1.6, 0.2])
dot_size = 3
font_size = 7
ress = pd.DataFrame({"cell_type": adata_sub.obs['celltype'].values,
"X1": adata_sub.obsm['X_tsne'][:,0],
"X2": adata_sub.obsm['X_tsne'][:,1]})
#fig, ax = plt.subplots(1, 5)
subfig_A0 = subfig_A[0].subfigures(1, 2, wspace=0.0, hspace=0, width_ratios=[4, 2])
ax = subfig_A0[0].subplots(1, 4)
sns.scatterplot(x='X1', y='X2', data=ress, alpha=0.9, s=dot_size,
linewidth=0, edgecolor="none", hue='cell_type',
ax=ax[0], legend='brief')
ax[0].axis('off')
ax[0].set_title("Cell types\n", fontsize=font_size)
ax[0].legend(bbox_to_anchor=[4.0, -0.01], ncol=3, prop={'size': font_size}, fontsize=font_size, frameon=False)
kwargs = dict(color='gray', density=.8, add_margin=.1, s=dot_size,
show=False, alpha=.2, min_mass=3.5, frameon=False)
scv.pl.velocity_embedding_stream(adata_sub, basis='tsne', fontsize=font_size, ax=ax[1], title='', **kwargs)
ax[1].set_title("Scvelo\n", fontsize=7)
scv.pl.velocity_embedding_stream(adata_sub, fontsize=font_size, basis='tsne',
title='', ax=ax[2], vkey='velocity_pyro', **kwargs)
ax[2].set_title("Pyro-Velocity\n", fontsize=7)
# plot_arrow_examples(adata_train, np.transpose(v_map_all, (1, 2, 0)), embeds_radian, ax=ax[3],
# n_sample=30, fig=fig, basis='umap', scale=0.004, alpha=0.18)
plot_arrow_examples(adata_sub, np.transpose(v_map_all, (1, 2, 0)), embeds_radian, ax=ax[3],
n_sample=30, fig=fig, basis='tsne', scale=0.001, alpha=0.3, index=100,
scale2=None, num_certain=0, num_total=4)
ax[3].set_title("Single cell\nvector field", fontsize=7)
plot_vector_field_uncertain(adata_sub, embed_mean, embeds_radian, fig=subfig_A0[1], cbar=True,
basis='tsne',
scale=0.018, arrow_size=5)
subfig_A0[0].subplots_adjust(hspace=0.2, wspace=0.1, left=0.01, right=0.99, top=0.99, bottom=0.45)
subfig_A0[1].subplots_adjust(hspace=0.2, wspace=0.1, left=0.01, right=0.99, top=0.99, bottom=0.45)
subfig_A[0].text(-0.06, 0.58, "PBMC", size=7, rotation="vertical", va="center")
V_grid.........
V_grid.........
60 50 40
[34]:
Text(-0.06, 0.58, 'PBMC')
[35]:
fig.savefig("fully_mature_pbmc_figure1.tif", facecolor=fig.get_facecolor(), bbox_inches='tight', edgecolor='none', dpi=300)
[42]:
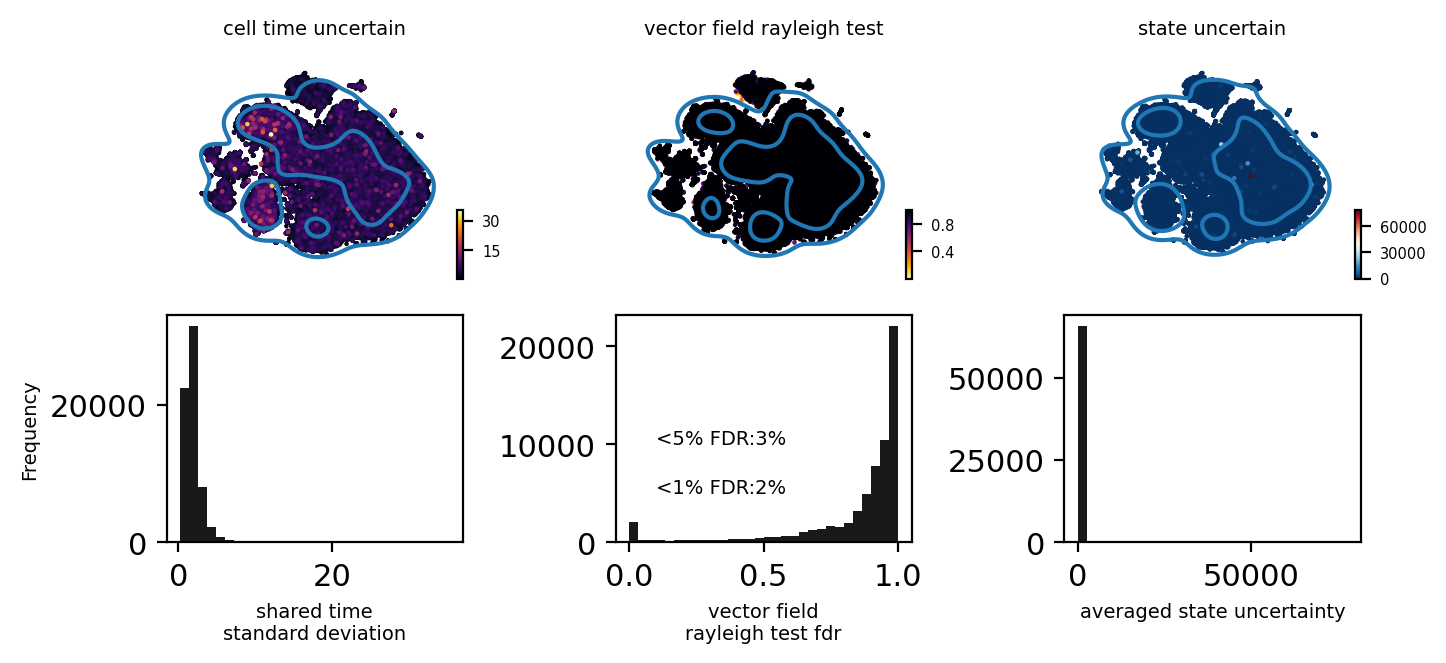
from pyrovelocity.plot import plot_state_uncertainty
fig, ax = plt.subplots(2, 3)
fig.set_size_inches(7.07, 3.5)
pos = adata_model_pos[1]
bin = 30
adata_sub.obs['cell_time_uncertain'] = adata_model_pos[1]['cell_time'].std(0).flatten()
scv.pl.scatter(adata_sub, c='cell_time_uncertain', ax=ax[0][0],
basis='tsne', show=False, cmap='inferno', fontsize=7)
select = adata_sub.obs['cell_time_uncertain'] > np.quantile(adata_sub.obs['cell_time_uncertain'], 0.9)
sns.kdeplot(adata_sub.obsm['X_tsne'][:, 0][select],
adata_sub.obsm['X_tsne'][:, 1][select], ax=ax[0][0], levels=3, fill=False)
_ = ax[1][0].hist(adata_sub.obs.cell_time_uncertain, bins=bin, color='black', alpha=0.9)
ax[1][0].set_xlabel("shared time\nstandard deviation", fontsize=7)
adata_sub.obs['vector_field_rayleigh_test'] = fdri
scv.pl.scatter(adata_sub, c='vector_field_rayleigh_test',
basis='tsne', ax=ax[0][1], show=False, cmap='inferno_r', fontsize=7)
select = adata_sub.obs['vector_field_rayleigh_test'] > np.quantile(adata_sub.obs['vector_field_rayleigh_test'], 0.9)
sns.kdeplot(adata_sub.obsm['X_tsne'][:, 0][select],
adata_sub.obsm['X_tsne'][:, 1][select], ax=ax[0][1], levels=3, fill=False)
_ = ax[1][1].hist(adata_sub.obs.vector_field_rayleigh_test, bins=bin, color='black', alpha=0.9)
ax[1][1].set_xlabel("vector field\nrayleigh test fdr", fontsize=7)
ax[1][1].text(0.1, 5e3, "<1%% FDR:%s%%" % int((fdri < 0.01).sum()/fdri.shape[0]*100), fontsize=7)
ax[1][1].text(0.1, 1e4, "<5%% FDR:%s%%" % int((fdri < 0.05).sum()/fdri.shape[0]*100), fontsize=7)
fig.subplots_adjust(hspace=0.3, wspace=0.7, left=0.01, right=0.8, top=0.92, bottom=0.17)
plot_state_uncertainty(pos, adata_sub,
basis='tsne', kde=True, data='raw', top_percentile=0.9, ax=ax[0][2])
_ = ax[1][2].hist(adata_sub.obs['state_uncertain'], bins=bin, color='black', alpha=0.9)
ax[1][2].set_xlabel("averaged state uncertainty", fontsize=7)
ax[1][0].set_ylabel("Frequency", fontsize=7)
fig.tight_layout()
[43]:
fig.savefig("fully_mature_pbmc_figure2.tif", facecolor=fig.get_facecolor(), bbox_inches='tight', edgecolor='none', dpi=300)
[ ]: