Lineage Tracing#
Load modules#
[1]:
%load_ext autoreload
%autoreload 2
import scvelo as scv
import numpy as np
import torch
import matplotlib.pyplot as plt
from pyrovelocity.data import load_data
from scipy.stats import spearmanr, pearsonr
from pyrovelocity.api import train_model
import seaborn as sns
import pandas as pd
from pyrovelocity.plot import plot_posterior_time, plot_gene_ranking,\
vector_field_uncertainty, plot_vector_field_uncertain,\
plot_mean_vector_field, project_grid_points,rainbowplot,denoised_umap,\
us_rainbowplot, plot_arrow_examples, set_colorbar
from pyrovelocity.utils import mae, mae_evaluate
from pyrovelocity.data import load_larry
Download filtered dataset#
see figshare
[2]:
adata_input = load_larry()
adata_input.layers['raw_spliced'] = adata_input.layers['spliced']
adata_input.layers['raw_unspliced'] = adata_input.layers['unspliced']
[3]:
scv.pp.filter_and_normalize(adata_input, min_shared_counts=30, n_top_genes=2000)
Filtered out 16583 genes that are detected 30 counts (shared).
Normalized count data: X, spliced, unspliced.
Extracted 2000 highly variable genes.
Logarithmized X.
[4]:
adata_input.obs['u_lib_size_raw'] = adata_input.layers['raw_unspliced'].toarray().sum(-1)
adata_input.obs['s_lib_size_raw'] = adata_input.layers['raw_spliced'].toarray().sum(-1)
[5]:
scv.pp.moments(adata_input, n_pcs=30, n_neighbors=30)
computing neighbors
finished (0:00:32) --> added
'distances' and 'connectivities', weighted adjacency matrices (adata.obsp)
computing moments based on connectivities
finished (0:00:05) --> added
'Ms' and 'Mu', moments of un/spliced abundances (adata.layers)
Apply Model 2 to all cells in LARRY (multi-fate dataset)#
[6]:
adata_model_pos = train_model(adata_input,
max_epochs=1000, svi_train=True, log_every=100,
patient_init=45,
batch_size=4000, use_gpu=0, cell_state='state_info',
include_prior=True,
offset=True,
library_size=True,
patient_improve=1e-3,
model_type='auto',
guide_type='auto',
train_size=1.0)
INFO No batch_key inputted, assuming all cells are same batch
INFO No label_key inputted, assuming all cells have same label
INFO Using data from adata.layers["raw_unspliced"]
INFO Using data from adata.layers["raw_spliced"]
INFO Successfully registered anndata object containing 49302 cells, 2000 vars, 1 batches, 1 labels, and 0
proteins. Also registered 0 extra categorical covariates and 0 extra continuous covariates.
INFO Please do not further modify adata until model is trained.
Anndata setup with scvi-tools version 0.13.0.
Data Summary ┏━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━┳━━━━━━━┓ ┃ Data ┃ Count ┃ ┡━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━╇━━━━━━━┩ │ Cells │ 49302 │ │ Vars │ 2000 │ │ Labels │ 1 │ │ Batches │ 1 │ │ Proteins │ 0 │ │ Extra Categorical Covariates │ 0 │ │ Extra Continuous Covariates │ 0 │ └──────────────────────────────┴───────┘
SCVI Data Registry ┏━━━━━━━━━━━━━━━━━━┳━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━┓ ┃ Data ┃ scvi-tools Location ┃ ┡━━━━━━━━━━━━━━━━━━╇━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━┩ │ X │ adata.layers['raw_spliced'] │ │ U │ adata.layers['raw_unspliced'] │ │ batch │ adata.obs['_scvi_batch'] │ │ label │ adata.obs['_scvi_labels'] │ │ ind_x │ adata.obs['_indices'] │ │ u_lib_size │ adata.obs['u_lib_size'] │ │ s_lib_size │ adata.obs['s_lib_size'] │ │ u_lib_size_mean │ adata.obs['u_lib_size_mean'] │ │ s_lib_size_mean │ adata.obs['s_lib_size_mean'] │ │ u_lib_size_scale │ adata.obs['u_lib_size_scale'] │ │ s_lib_size_scale │ adata.obs['s_lib_size_scale'] │ └──────────────────┴───────────────────────────────┘
Label Categories ┏━━━━━━━━━━━━━━━━━━━━━━━━━━━┳━━━━━━━━━━━━┳━━━━━━━━━━━━━━━━━━━━━┓ ┃ Source Location ┃ Categories ┃ scvi-tools Encoding ┃ ┡━━━━━━━━━━━━━━━━━━━━━━━━━━━╇━━━━━━━━━━━━╇━━━━━━━━━━━━━━━━━━━━━┩ │ adata.obs['_scvi_labels'] │ 0 │ 0 │ └───────────────────────────┴────────────┴─────────────────────┘
Batch Categories ┏━━━━━━━━━━━━━━━━━━━━━━━━━━┳━━━━━━━━━━━━┳━━━━━━━━━━━━━━━━━━━━━┓ ┃ Source Location ┃ Categories ┃ scvi-tools Encoding ┃ ┡━━━━━━━━━━━━━━━━━━━━━━━━━━╇━━━━━━━━━━━━╇━━━━━━━━━━━━━━━━━━━━━┩ │ adata.obs['_scvi_batch'] │ 0 │ 0 │ └──────────────────────────┴────────────┴─────────────────────┘
-----------
auto
auto
step 0 loss = 2.64038e+08 patience = 45
step 100 loss = 1.83999e+08 patience = 45
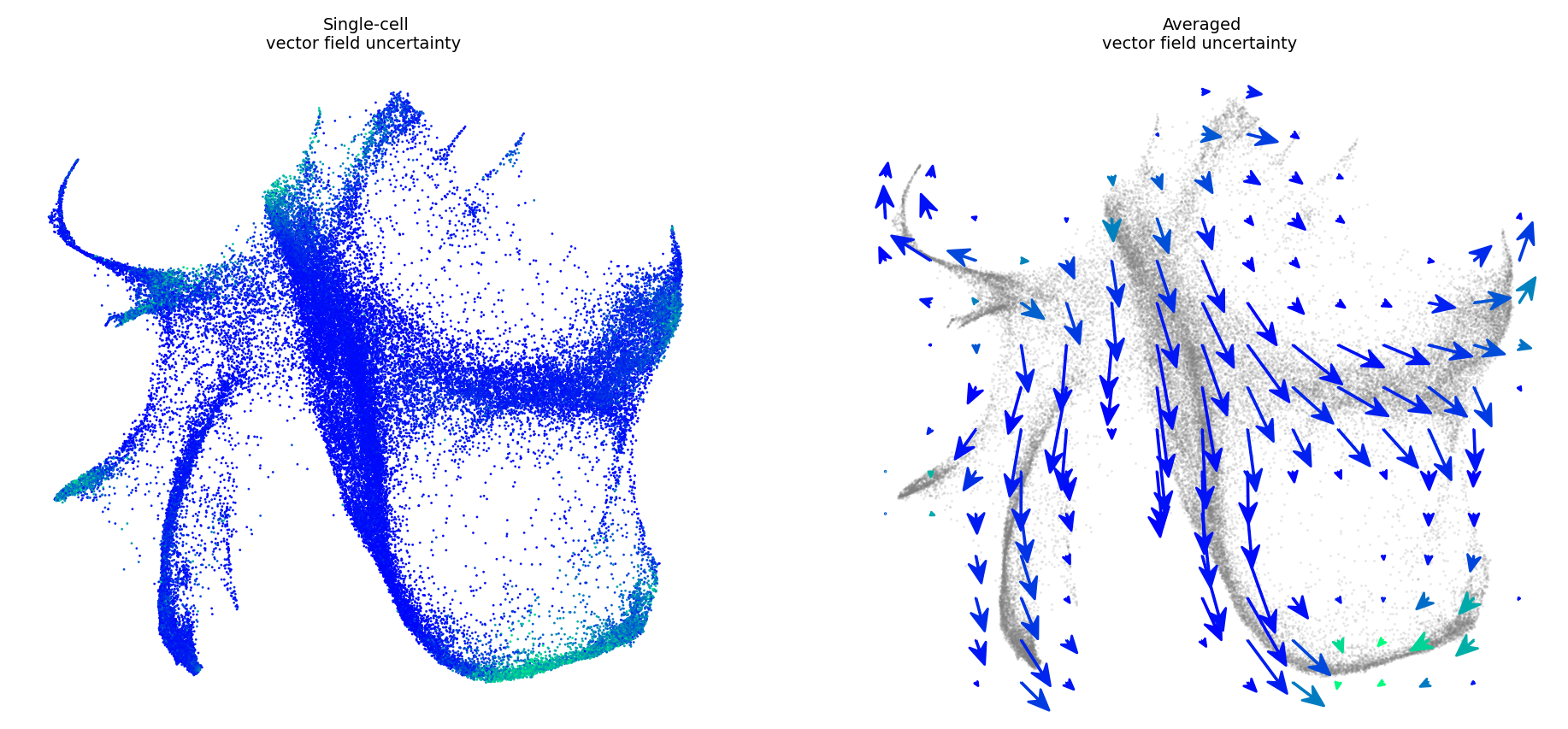
[7]:
v_map_all, embeds_radian, fdri = vector_field_uncertainty(adata_input, adata_model_pos[1],
basis='emb', denoised=False, n_jobs=30)
computing neighbors
finished (0:00:12) --> added
'distances' and 'connectivities', weighted adjacency matrices (adata.obsp)
computing velocity graph (using 30/256 cores)
finished (0:02:24) --> added
'velocity_pyro_graph', sparse matrix with cosine correlations (adata.uns)
computing velocity embedding
finished (0:00:10) --> added
'velocity_pyro_emb', embedded velocity vectors (adata.obsm)
computing velocity graph (using 30/256 cores)
finished (0:01:46) --> added
'velocity_pyro_graph', sparse matrix with cosine correlations (adata.uns)
computing velocity embedding
finished (0:00:10) --> added
'velocity_pyro_emb', embedded velocity vectors (adata.obsm)
computing velocity graph (using 30/256 cores)
finished (0:01:46) --> added
'velocity_pyro_graph', sparse matrix with cosine correlations (adata.uns)
computing velocity embedding
finished (0:00:11) --> added
'velocity_pyro_emb', embedded velocity vectors (adata.obsm)
computing velocity graph (using 30/256 cores)
finished (0:01:45) --> added
'velocity_pyro_graph', sparse matrix with cosine correlations (adata.uns)
computing velocity embedding
finished (0:00:11) --> added
'velocity_pyro_emb', embedded velocity vectors (adata.obsm)
computing velocity graph (using 30/256 cores)
finished (0:01:47) --> added
'velocity_pyro_graph', sparse matrix with cosine correlations (adata.uns)
computing velocity embedding
finished (0:00:10) --> added
'velocity_pyro_emb', embedded velocity vectors (adata.obsm)
computing velocity graph (using 30/256 cores)
finished (0:01:44) --> added
'velocity_pyro_graph', sparse matrix with cosine correlations (adata.uns)
computing velocity embedding
finished (0:00:11) --> added
'velocity_pyro_emb', embedded velocity vectors (adata.obsm)
computing velocity graph (using 30/256 cores)
finished (0:01:44) --> added
'velocity_pyro_graph', sparse matrix with cosine correlations (adata.uns)
computing velocity embedding
finished (0:00:11) --> added
'velocity_pyro_emb', embedded velocity vectors (adata.obsm)
computing velocity graph (using 30/256 cores)
finished (0:01:45) --> added
'velocity_pyro_graph', sparse matrix with cosine correlations (adata.uns)
computing velocity embedding
finished (0:00:10) --> added
'velocity_pyro_emb', embedded velocity vectors (adata.obsm)
computing velocity graph (using 30/256 cores)
finished (0:01:46) --> added
'velocity_pyro_graph', sparse matrix with cosine correlations (adata.uns)
computing velocity embedding
finished (0:00:10) --> added
'velocity_pyro_emb', embedded velocity vectors (adata.obsm)
computing velocity graph (using 30/256 cores)
finished (0:01:45) --> added
'velocity_pyro_graph', sparse matrix with cosine correlations (adata.uns)
computing velocity embedding
finished (0:00:11) --> added
'velocity_pyro_emb', embedded velocity vectors (adata.obsm)
computing velocity graph (using 30/256 cores)
finished (0:01:46) --> added
'velocity_pyro_graph', sparse matrix with cosine correlations (adata.uns)
computing velocity embedding
finished (0:00:11) --> added
'velocity_pyro_emb', embedded velocity vectors (adata.obsm)
computing velocity graph (using 30/256 cores)
finished (0:01:47) --> added
'velocity_pyro_graph', sparse matrix with cosine correlations (adata.uns)
computing velocity embedding
finished (0:00:10) --> added
'velocity_pyro_emb', embedded velocity vectors (adata.obsm)
computing velocity graph (using 30/256 cores)
finished (0:01:45) --> added
'velocity_pyro_graph', sparse matrix with cosine correlations (adata.uns)
computing velocity embedding
finished (0:00:10) --> added
'velocity_pyro_emb', embedded velocity vectors (adata.obsm)
computing velocity graph (using 30/256 cores)
finished (0:01:45) --> added
'velocity_pyro_graph', sparse matrix with cosine correlations (adata.uns)
computing velocity embedding
finished (0:00:11) --> added
'velocity_pyro_emb', embedded velocity vectors (adata.obsm)
computing velocity graph (using 30/256 cores)
finished (0:01:43) --> added
'velocity_pyro_graph', sparse matrix with cosine correlations (adata.uns)
computing velocity embedding
finished (0:00:11) --> added
'velocity_pyro_emb', embedded velocity vectors (adata.obsm)
computing velocity graph (using 30/256 cores)
finished (0:01:44) --> added
'velocity_pyro_graph', sparse matrix with cosine correlations (adata.uns)
computing velocity embedding
finished (0:00:11) --> added
'velocity_pyro_emb', embedded velocity vectors (adata.obsm)
computing velocity graph (using 30/256 cores)
finished (0:01:43) --> added
'velocity_pyro_graph', sparse matrix with cosine correlations (adata.uns)
computing velocity embedding
finished (0:00:10) --> added
'velocity_pyro_emb', embedded velocity vectors (adata.obsm)
computing velocity graph (using 30/256 cores)
finished (0:01:45) --> added
'velocity_pyro_graph', sparse matrix with cosine correlations (adata.uns)
computing velocity embedding
finished (0:00:10) --> added
'velocity_pyro_emb', embedded velocity vectors (adata.obsm)
computing velocity graph (using 30/256 cores)
finished (0:01:45) --> added
'velocity_pyro_graph', sparse matrix with cosine correlations (adata.uns)
computing velocity embedding
finished (0:00:10) --> added
'velocity_pyro_emb', embedded velocity vectors (adata.obsm)
computing velocity graph (using 30/256 cores)
finished (0:01:45) --> added
'velocity_pyro_graph', sparse matrix with cosine correlations (adata.uns)
computing velocity embedding
finished (0:00:11) --> added
'velocity_pyro_emb', embedded velocity vectors (adata.obsm)
computing velocity graph (using 30/256 cores)
finished (0:01:47) --> added
'velocity_pyro_graph', sparse matrix with cosine correlations (adata.uns)
computing velocity embedding
finished (0:00:11) --> added
'velocity_pyro_emb', embedded velocity vectors (adata.obsm)
computing velocity graph (using 30/256 cores)
finished (0:01:46) --> added
'velocity_pyro_graph', sparse matrix with cosine correlations (adata.uns)
computing velocity embedding
finished (0:00:10) --> added
'velocity_pyro_emb', embedded velocity vectors (adata.obsm)
computing velocity graph (using 30/256 cores)
finished (0:01:45) --> added
'velocity_pyro_graph', sparse matrix with cosine correlations (adata.uns)
computing velocity embedding
finished (0:00:10) --> added
'velocity_pyro_emb', embedded velocity vectors (adata.obsm)
computing velocity graph (using 30/256 cores)
finished (0:01:46) --> added
'velocity_pyro_graph', sparse matrix with cosine correlations (adata.uns)
computing velocity embedding
finished (0:00:10) --> added
'velocity_pyro_emb', embedded velocity vectors (adata.obsm)
computing velocity graph (using 30/256 cores)
finished (0:01:44) --> added
'velocity_pyro_graph', sparse matrix with cosine correlations (adata.uns)
computing velocity embedding
finished (0:00:10) --> added
'velocity_pyro_emb', embedded velocity vectors (adata.obsm)
computing velocity graph (using 30/256 cores)
finished (0:01:45) --> added
'velocity_pyro_graph', sparse matrix with cosine correlations (adata.uns)
computing velocity embedding
finished (0:00:10) --> added
'velocity_pyro_emb', embedded velocity vectors (adata.obsm)
computing velocity graph (using 30/256 cores)
finished (0:01:46) --> added
'velocity_pyro_graph', sparse matrix with cosine correlations (adata.uns)
computing velocity embedding
finished (0:00:10) --> added
'velocity_pyro_emb', embedded velocity vectors (adata.obsm)
computing velocity graph (using 30/256 cores)
finished (0:01:43) --> added
'velocity_pyro_graph', sparse matrix with cosine correlations (adata.uns)
computing velocity embedding
finished (0:00:11) --> added
'velocity_pyro_emb', embedded velocity vectors (adata.obsm)
computing velocity graph (using 30/256 cores)
finished (0:01:45) --> added
'velocity_pyro_graph', sparse matrix with cosine correlations (adata.uns)
computing velocity embedding
finished (0:00:11) --> added
'velocity_pyro_emb', embedded velocity vectors (adata.obsm)
computing velocity graph (using 30/256 cores)
finished (0:01:45) --> added
'velocity_pyro_graph', sparse matrix with cosine correlations (adata.uns)
computing velocity embedding
finished (0:00:10) --> added
'velocity_pyro_emb', embedded velocity vectors (adata.obsm)
[8]:
fig, ax = plt.subplots()
embed_mean = plot_mean_vector_field(adata_model_pos[1], adata_input, ax=ax, n_jobs=30, basis='emb')
computing neighbors
finished (0:00:12) --> added
'distances' and 'connectivities', weighted adjacency matrices (adata.obsp)
computing velocity graph (using 30/256 cores)
finished (0:01:46) --> added
'velocity_pyro_graph', sparse matrix with cosine correlations (adata.uns)
computing velocity embedding
finished (0:00:11) --> added
'velocity_pyro_emb', embedded velocity vectors (adata.obsm)

[9]:
fig, ax = plt.subplots(1, 2)
fig.set_size_inches(11.5, 5)
plot_vector_field_uncertain(adata_input, embed_mean, embeds_radian,
ax=ax,
fig=fig, cbar=False, basis='emb', scale=None)
V_grid.........
60 50 40
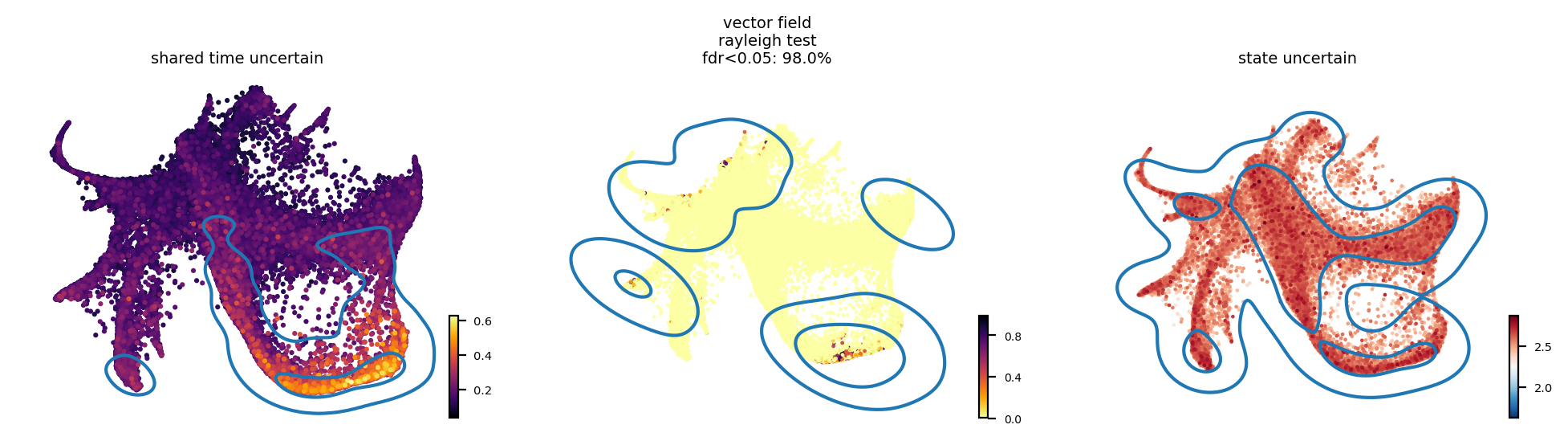
More uncertainty estimation for LARRY#
[10]:
from pyrovelocity.plot import plot_state_uncertainty
fig, ax = plt.subplots(1, 3)
fig.set_size_inches(12, 2.8)
adata_input.obs['shared_time_uncertain'] = adata_model_pos[1]['cell_time'].std(0).flatten()
ax_cb = scv.pl.scatter(adata_input, c='shared_time_uncertain', ax=ax[0], show=False, cmap='inferno', fontsize=7, s=20, colorbar=True, basis='emb')
select = adata_input.obs['shared_time_uncertain'] > np.quantile(adata_input.obs['shared_time_uncertain'], 0.9)
sns.kdeplot(adata_input.obsm['X_emb'][:, 0][select],
adata_input.obsm['X_emb'][:, 1][select],
ax=ax[0], levels=3, fill=False)
adata_input.obs.loc[:, 'vector_field_rayleigh_test'] = fdri
basis = 'emb'
im = ax[1].scatter(adata_input.obsm[f'X_{basis}'][:, 0],
adata_input.obsm[f'X_{basis}'][:, 1], s=3, alpha=0.9,
c=adata_input.obs['vector_field_rayleigh_test'], cmap='inferno_r',
linewidth=0)
##ax[1].set_title("Rayleigh test")
set_colorbar(im, ax[1], labelsize=5, fig=fig, position='right')
select = adata_input.obs['vector_field_rayleigh_test'] > np.quantile(adata_input.obs['vector_field_rayleigh_test'], 0.95)
sns.kdeplot(adata_input.obsm['X_emb'][:, 0][select],
adata_input.obsm['X_emb'][:, 1][select], ax=ax[1], levels=3, fill=False)
ax[1].axis('off')
ax[1].set_title("vector field\nrayleigh test\nfdr<0.05: %s%%" % (round((fdri < 0.05).sum()/fdri.shape[0], 2)*100), fontsize=7)
_ = plot_state_uncertainty(adata_model_pos[1], adata_input, kde=True, data='raw',
top_percentile=0.9, ax=ax[2], basis='emb')
/PHShome/qq06/ssd/alvin/conda/envs/pyrovelocity_anaconda/lib/python3.8/site-packages/scvelo/plotting/utils.py:869: MatplotlibDeprecationWarning: The draw_all function was deprecated in Matplotlib 3.6 and will be removed two minor releases later. Use fig.draw_without_rendering() instead.
cb.draw_all()
/PHShome/qq06/ssd/alvin/conda/envs/pyrovelocity_anaconda/lib/python3.8/site-packages/pyrovelocity/plot.py:1116: MatplotlibDeprecationWarning: The draw_all function was deprecated in Matplotlib 3.6 and will be removed two minor releases later. Use fig.draw_without_rendering() instead.
cb.draw_all()
/PHShome/qq06/ssd/alvin/conda/envs/pyrovelocity_anaconda/lib/python3.8/site-packages/scvelo/plotting/utils.py:869: MatplotlibDeprecationWarning: The draw_all function was deprecated in Matplotlib 3.6 and will be removed two minor releases later. Use fig.draw_without_rendering() instead.
cb.draw_all()